Followers of these pages will know that over the last decade our picture of solid tumours has gradually resolved into an ecosystem comprising a wide variety of cells, including numerous different types of immune cell, blood vessels and the extracellular matrix. In addition tumour-associated microbiota — microorganisms, predominantly bacteria — have emerged as an intrinsic part of the ‘tumour microenvironment’ in many cancer types.
The story so far
In the human gut there are about 2,000 different species of bacteria that, together, carry many hundreds more genes than the 20,000 encoded in the human genome. We’ve seen that that obesity causes a switch in the proportions of two major sub-families of bacteria, resulting in a decrease in the number of bug genes (it’s a small world). Even more remarkably, there’s evidence that the microbiome can affect metastasis (Hitchhiker Or Driver?).
Drawing the map
Now, in a really ambitious effort, Jorge Luis Galeano Niño, Susan Bullman and colleagues from the Fred Hutchinson Cancer Center, Seattle and other US centres have applied in situ spatial-profiling technologies and single-cell RNA sequencing to two types of human tumours. The idea was to map the presence of bacteria across the tumours. They used 77 antibodies to detect protein markers of particular cell types and major cancer-associated signalling pathways. On top of that they developed a single-cell RNA transcript profiling method that sequences human transcripts alongside bacterial rDNA transcripts in the same cell.
What did they find?
As ever, that bald summary does no justice to the technical brilliance involved but, as ever, what we’re really interested in are the results. A key finding was that Fusobacterium nucleatum is one of the most abundant types of bacterium in oral squamous cell carcinoma and colorectal cancer, consistent with previous work (Hitchhiker Or Driver?). Proteins associated with immunosuppression (PD-1 and CTLA4), were highly expressed in bacteria-rich areas of tumours, whereas hallmarks of cell proliferation (Ki67), were depleted, a further indication of a reduced immune response. This indicates what has been called a ‘proliferation–migration trade-off’ in which there is increased expression of genes associated with cell migration but inhibition of proliferation-associated and DNA repair genes.
Notably, bacteria were more common in tumour cells with an abnormal, rather than a normal, number of chromosomes, suggesting a bacterial preference for cancer cells over normal cells.
Finally, and very excitingly, colorectal cancer cells grown in a 3D system in the lab had an increased migration capacity when infected with Fusobacterium.
Features of bacteria in a tumour. Regions low in bacteria were associated with the presence of more blood vessels, and had high levels of T cells and low levels of myeloid cells. Cancer cells in these regions had higher levels of proliferation. For regions with high bacterial levels (inside tumour and myeloid cells) myeloid cells were more common and T cells were rare and showed signs of immunosuppression — expression of the proteins PD-1 and CTLA4. Cancer cells in such regions also had a higher capacity for migration. From Livyatan and Straussman, 2022.
Bacteria stick to and enter oral squamous cell carcinoma tumour cells. Confocal images of bacteria-associated single cells after tissue dissociation. From Niño et al., 2022.
The spatial distribution of intra-tumoral bacteria in human tumours. The pixels are total bacterial reads: red = highest level. Left: oral squamous cell carcinoma. Right: colorectal cancer. From Niño et al., 2022.
Niño et al. mapped the 10 most common species in the two types of tumour. These varied between tumours and only Fusobacterium was present in a significant fraction of both tumours. Moreover, the distribution of microbiota within a tumour is not random (see image above) but is highly organised in microniches together with host cells that promote aggression.
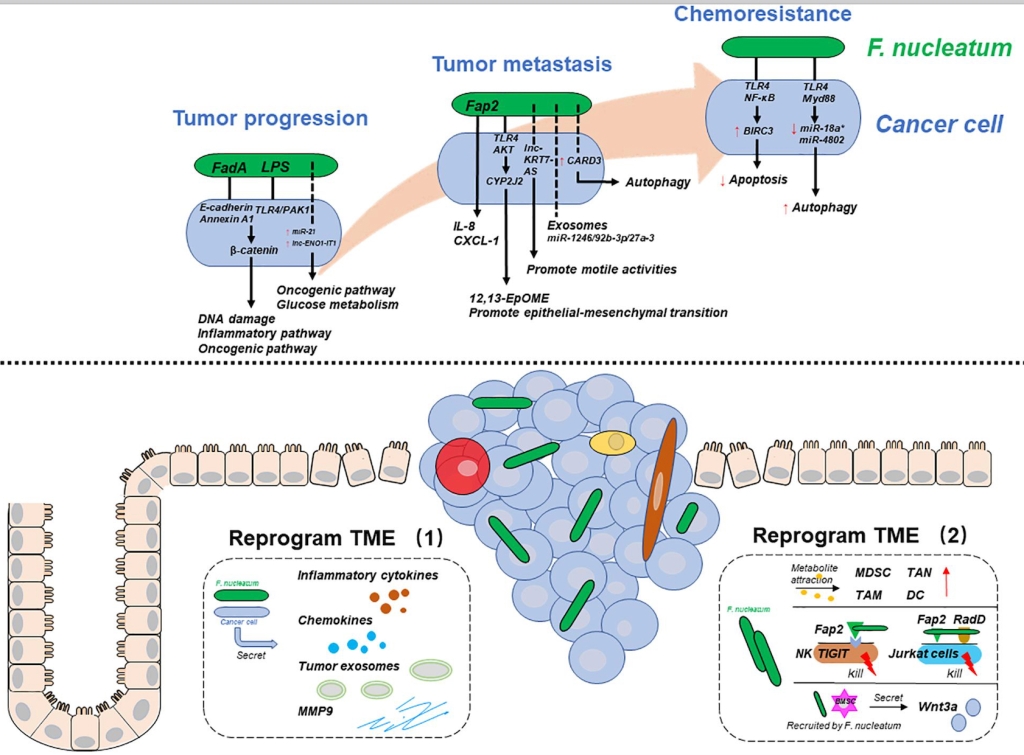
Fusobacterium and colorectal cancer. (A) Fusobacterium reprograms cancer cells during tumour progression, metastasis, and chemoresistance. (B) Fusobacterium reprograms the tumour microenvironment. From Wang et al., 2021.
The details of how F. nucleatum exerts its effects are still far from clear but there are some pointers. FadA and Fap2 permit bacteria to stick to cells and LPS regulates the inflammation signal through the innate immune pathway.
For the moment we should salute a technical tour de force that is not perfect in design nor complete in its answers but gives us the clearest picture yet of the complexity of the tumour microenvironment and the astonishing effects that can be wrought by our tiny co-dwellers.
References
Galeano Niño, J.L., Wu, H., LaCourse, K.D. et al. Effect of the intratumoral microbiota on spatial and cellular heterogeneity in cancer. Nature (2022). https://doi.org/10.1038/s41586-022-05435-0.
Livyatan, I. and Ravid Straussman, R. (2022). A spatial perspective on bacteria in tumours. Nature 16 November 2022. https://doi.org/10.1038/d41586-022-03669-6.
Wang S, Liu Y, Li J, Zhao L, Yan W, Lin B, Guo X and Wei Y (2021). Fusobacterium nucleatum Acts as a Pro-carcinogenic Bacterium in Colorectal Cancer: From Association to Causality. Front. Cell Dev. Biol. 9:710165. doi: 10.3389/fcell.2021.710165.