It hardly needs saying that these days advances in medicine, and indeed in all science, are technology driven. That is, significant steps need help in the form of new or improved methods. No genome sequences without Fred Sanger’s dideoxy nucleotide chain terminators. Often these technical steps go largely unremarked but once in a while they hit the field like a seismic event — Fred’s terminators. And I guess if you asked a random sample of molecular biologists for the Mega event of the last decade they’d probably go for CRISPR-Cas9 genome editing developed by Emmanuelle Charpentier and Jennifer Doudna. Its spectacular impact was due to that fact that it permitted gene editing within organisms — a feat that had hitherto seemed in the realms of science fiction. It was indeed a wondrous achievement and in just a few months practically every molecular biology lab on the planet was giving it a go.
A dream comes true
And it worked too! It wasn’t long before CRISPR was having stunning effects in cancer patients (Precision CAR-T steering). Of course, nothing’s perfect and it was predictable that CRISPR would turn out to be less than 100% efficient, to have off-target effects and, when it cuts DNA, to sometimes have drastic effects on the cell (Caveat Emptor).
Helpfully in those two blogs we explained that CRISPR (clustered regularly interspaced short palindromic repeats) is a family of DNA sequences in prokaryotes (e.g., bacteria) that come from things (bacteriophages) that had previously infected the bug. They play a key role in defending it against subsequent infections. Modifying this system for gene editing uses the enzyme Cas9 (CRISPR-associated protein 9) to target CRISPR sequences as a guide to recognize specific regions of DNA complementary to the CRISPR sequence. The enzyme then makes double strand breaks in DNA that are repaired by the machinery of the cell, a step that enables existing genes to be removed and/or new ones added in vivo. Specific targetting is achieved by delivering the Cas9 nuclease complexed with a synthetic guide RNA (gRNA) into the cell.
Destroying the evidence
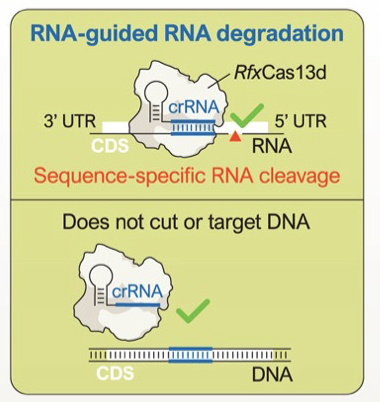
So far so brilliant but now Stanley Qi, Crystal Mackall and colleagues at Stanford University in California have come up with an alternative system to get round the problems of CRISPR-Cas9. Called MEGA (multiplexed effector guide arrays) it still uses a CRISPR guide RNA but swaps the DNA-cutting Cas9 for an RNA-cutting alternative called Cas13d. Thus the CRISPR bit directs Cas13d to a target mRNA (produced from a DNA template) rather than to DNA.
Editing the transcriptome (RNA) in human T cells. MEGA (multiplexed effector guide arrays) use a CRISPR guide RNA but the cutting enzyme is Cas13d that cuts RNA not DNA. From Tieu et al., 2024.
Messenger RNA (mRNA) is the information carrier between DNA and protein — hence the idea behind MEGA is that by targeting mRNAs in dysfunctional CAR-T cells the encoded proteins won’t be made, tumour cell killing will be restored and the T cell genome (DNA) will be unaffected.
Nerds corner: How they did it
Primary human T cells were transduced with combinations of lentiviruses (retroviruses used to introduce new genes to cells) to deliver Cas 13d and, in separate lentiviruses, to deliver RNA guides targeting RNAs that are over-expressed CAR-T cells. By nicking bits of virus or bacterial genomes, it’s possible not only to make such inserts switchable (on or off in terms of expression) but to adjust (titrate) their activity. Stanley Qi & Co targeted three genes (LAG3, PD-1 and TIM3) that are typically upregulated in dysfunctional CAR-T cells. Sure enough, they were able to block completely (knock-down) expression of these genes but also to regulate their level of expression — i.e. to give ‘tuneable’ regulation of mRNA (and hence) protein levels.
All told, a remarkable demonstration of the power of modern molecular biology.
And the results …
From all this complexity it emerged that knock-down of more than one gene was more effective than hitting a single gene in terms of T cell proliferation and tumour cell killing. They were able to knock down groups of up to 10 genes by this method, illustrating an enormous potential that goes beyond even improving anti-tumour activity.
References
Tieu, V. et al. 2024. A versatile CRISPR-Cas13d platform for multiplexed transcriptomic regulation and metabolic engineering in primary human T cells. Cell 187, 2024, 1278-1295.e20. https://doi.org/10.1016/j.cell.2024.01.035.