Readers of this blog will have noted frequent references to The Human Genome Project — that produced the first complete sequence of human DNA — and how subsequent amazing technical advances now mean that genomes can be sequenced on an industrial scale. This has led to the Pan-Cancer Atlas, a global, collaborative effort that has analysed over 11,000 tumours of the 33 most prevalent cancers that we explained in Be amazed and No It Isn’t!
Not wishing to be left behind we need to grasp the thrust of the most recent step in this saga — a mutational signature analysis of just over 12,000 whole genome sequences of tumour matched with normal sequence from corresponding tissues. The work was done by Andrea Degasperi, Serena Nik-Zainal and colleagues at Cambridge University and they also added a couple of other studies to their analysis giving almost 19,000 sequenced cancers in total. Analysing this vast amount of data they identified single-base substitution (SBS) and double-base substitution (DBS) signatures independently in each organ.
Never mind the data!
All who keep even an occasional eye on cancer molecular biology will have a picture of what’s in this paper: an absolutely mind-blowing amount of data! So let’s put that to one side, together with the technicalities of how it was done (sorry Andrea!) and just tease out the upshot.
The analysis focussed on identifying SBSs and DBSs as signatures. A single base substitution (aka single nucleotide variant) is when one nucleotide base in DNA is changed to another (the possible substitutions are: C>A, C>G, C>T, T>A, T>C, and T>G). Double base substitutions swap two adjacent DNA base-pairs (e.g., CT:GA to AA:TT, often written as CT:GA > AA:TT).

An example of a single-base substitution signature, SBS4 (click to view axes). SBS4 is a tobacco-associated signature with a high frequency of C>A mutations occurring mainly in lung cancers (in this case a high frequency means about 90 substitutions per megabase of DNA. SBS4 occurs very rarely in other tumour types (e.g., observed in one bladder cancer, one breast cancer, one metastatic bowel cancer one astrocytoma and three central nervous system tumours) with the exception of liver to which lung tumours commonly spread (86% of liver samples carried SBS4). The y-axis is the probability of finding the mutation; the x-axis shows the 16 possible triplet sequences containing a C>A mutation. All the identified SBS signatures can be viewed together with their frequency in tumour types at: https://signal.mutationalsignatures.com/explore/study/6?mutationType=1. From Degasperi et al. 2022.
The aim was to produce a kind of family tree of each individual cancer — how damage to DNA (mutations) and the repair processes that have gone on yield a ‘mutational signature’ — a cumulative picture of how external and internal (environmental and endogenous) factors have come together to cause each cancer. And the grand idea is to show that it is realistic to determine all the key ‘driver’ mutations in an individual cancer — a critical step to the goal of ‘personalized medicine’.
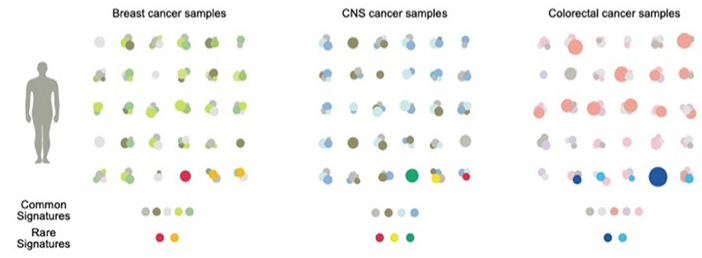
Patterns of mutational signatures in breast, central nervous system (CNS) and colorectal cancers. Common signatures are shown in gray and lighter colours. Rare signatures in red, yellow and dark and light blue. From Degasperi et al. 2022.
This study developed a new analytical package designed to fit common and rare signatures as a step to personalized therapy. The patterns revealed that each tumour sample may have different amounts of some or all of the common signatures. They may also carry a rare signature (examples in the bottom line of the figure above). Common signatures are a mixture: some occur in almost all types of tumour whilst others show restricted occurrence. Rare signatures may be unique, i.e. confined to one type of tumour, but some occur across tumour types (e.g., red dots). Collecting this type of data is important, not least because identifying the same mutation signature in tumours in different tissues will lead to treatments based on genetics — a concept that is gradually displacing the time-honoured approach of classifying cancers by tissue of origin.
Reference
Degasperi, A, Zou X, Amarante TD, Martinez-Martinez A, Koh GCC, Dias JML, Heskin L, Chmelova L, Rinaldi G, Wang VYW, Nanda AS, Bernstein A, Momen SE, Young J, Perez-Gil D, Memari Y, Badja C, Shooter S, Czarnecki J, Brown MA, Davies HR. (2022). Genomics England Research Consortium, Nik-Zainal S. Substitution mutational signatures in whole-genome-sequenced cancers in the UK population. Science 376(6591):science.abl9283. doi: 10.1126/science.abl9283. PMID: 35949260; PMCID: PMC7613262.