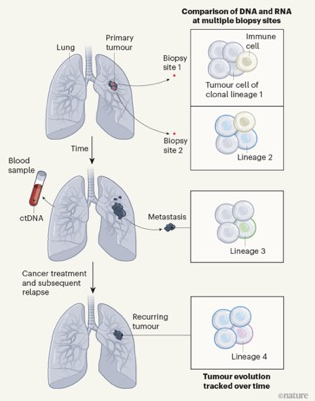
In the previous blog we caught up with the ongoing lung TRACERx study, specifically its work on the effect of air-born pollutants. However, TRACERx has a much broader overall aim, namely to follow the evolution of human lung cancers over time — tracking patterns of cell lineages (clones) of tumour cells, monitoring gene expression, tracking circulating tumour DNA (ctDNA) in the bloodstream and defining how immune cells target lung cancer (see figure below).
It’s important because lung cancer is the leading global cause of cancer-related deaths. The two subtypes are non-small cell lung cancer (NSCLC: 85% of cases), and small cell lung cancer (the remaining 15%). NSCLC comes in three subtypes based on what the cells look like (lung adenocarcinoma, lung squamous cell carcinoma and large cell carcinoma).
The scale of these studies is hard to grasp for they examined NSCLC in 421 individuals and did genomic profiles of 1,644 tumour regions.
The moving picture of lung cancer. The scheme outlines the TRACERx project: tumour samples are analysed for patterns of cell lineages (clones) by comparing DNA and RNA sequences, for immune cells in the tumour microenvironment and for changes in tumour cells that have spread (metastasized) to distant sites. Circulating tumour DNA (ctDNA) shed into the bloodstream was also screened to monitor disease progression and response to treatment. From Hayes and Meyerson 2023.
The main aim
The central idea behind TRACERx is to map how the variables summarised above relate to clinical outcome — i.e. disease-free survival, the survival time after treatment without cancer symptoms. The approach is to get a view of the constantly changing picture of tumour heterogeneity by sequencing the protein-coding regions of known cancer driving genes (by whole-exome sequencing or by sequencing the whole-genome). This should reveal the effects of chemotherapy (e.g., platinum-based drugs).
In addition to the air-born pollutant work, the consortium has just published four more papers that reflect progress towards these aims. For the purposes of this blog we will attempt a brief summary of these — with sincere apologies to those whose fantastic work has thus been subsumed.
Jargon buster
First a note about the terminology of mutations in cancer:
(i) clonal mutations are shared by all cancer cells.
(ii) subclonal mutations are present only in a subset of tumour cells. A subclone is a descendant of the most recent common ancestor (MRCA) of the tumour sample.
(iii) truncal mutations are mutations present in the trunk of the cancer evolutionary tree, i.e. mutations present in all subclones and at all timepoints.
(iv) Whole-genome doubling (WGD) involves doubling of the entire chromosome complement. It is a prevalent event in cancer.
(v) A clonal sweep occurs when a subclone outcompetes its neighboring cells, resulting in a trend of reduced diversity and towards a homogeneous tumour.
Key points
1. Lung adenocarcinoma: Frankell et al. found sub-clonal mutations in 22 of 40 common ‘cancer genes’. These included TP53 and KRAS. Truncal mutations in TP53 and KRAS tended to be mutually exclusive and these were also exclusive for EGFR whole genome doubling (WGD). Amplification of MYC and activating mutations in receptor tyrosine kinase pathways (both major cancer drivers) were truncal events, in contrast to the generally subclonal events affecting TP53 and KRAS, despite the latter being major cancer drivers.
Subclonal WGD was detected in 19% of tumours and 10% of tumours had multiple subclonal WGDs. Subclonal, but not truncal, WGD was associated with shorter disease-free survival. Notably, 8% of these tumours in smokers showed no evidence of tobacco-induced mutations but they carried patterns of mutations in EGFR and other oncogenes (RET, ROS1, ALK and MET) resembling those found in never-smokers, suggesting similar causes.
In 1% of cases lung tumours were observed that had two distinct genomic origins. Called ‘collision tumours’ these consist of two distinct tumours that have grown to occupy the same region of the lung as a single, continuous mass.
Overall these results show the importance of clonal expansion, WGD and copy number instability in regulating the behaviour of non-small cell lung cancer.
2. Metastasis: Al Bakir et al. compared the genomes (DNA sequences) of primary tumours with those of their metastases and found that in 25% of cases metastatic growth diverged early, before the final clonal sweep in the primary. This shows up as a set of mutations in all regions of the primary that was absent from metastases. Early divergence often occurred in small tumours (less than 8 mm diameter) and was more common in smokers. All of which highlights the current limitations of radiological screening for identifying early diverging tumours and the problems of targeting metastasis-seeding subclones.
3. Metastasis: Abbosh et al. developed a bioinformatics method (ECLIPSE) to track low levels of ctDNA and thus identify cases of polyclonal metastatic dissemination that are associated with poor outcome. They also showed that ctDNA could forecast impending relapse and that it is useful for selecting patients who might benefit from drug treatments.
4. Intra-tumour heterogeneity: Martínez-Ruiz et al. looked at gene expression (i.e. the RNA molecules being made {transcribed} at any time) in non-small cell lung tumours and found that the outgrowth of a clone containing a specific mutation was positively selected when that gene was highly expressed. In addition they looked at different versions (alleles) of the same gene and found that both copy-number-dependent and copy-number-independent events could affect the expression of an allele (gene copy number means the number of copies of a given gene in an organism’s complete set of genes).
It emerged that copy-number-independent events affected epigenetic regulators (i.e. DNA and histone modulators). In particular mutations in a set of epigenetic modifiers (CREBBP, KDM5C, SMARCA4, SETD2 and KMT2B) were associated with increased levels of copy-number-independent alleles. By contrast, mutations in the de-methylase KDM6A were associated with decreased expression of these alleles.
These summaries do no justice at all to the huge amount of work that has produced these papers. Far from answering all the problems of lung cancer, they rather highlight our ignorance — but the advances they make on a range of fronts show that the enigma of lung cancer is slowly being prized open.
References
Hayes TK, Meyerson M. Molecular portraits of lung cancer evolution. Nature. 2023 Apr;616(7957):435-436. doi: 10.1038/d41586-023-00934-0. PMID: 37045956.
Frankell, A.M., Dietzen, M., Al Bakir, M. et al. The evolution of lung cancer and impact of subclonal selection in TRACERx. Nature 616, 525–533 (2023). https://doi.org/10.1038/s41586-023-05783-5
Al Bakir, M., Huebner, A., Martínez-Ruiz, C. et al. The evolution of non-small cell lung cancer metastases in TRACERx. Nature 616, 534–542 (2023). https://doi.org/10.1038/s41586-023-05729-x
Abbosh, C., Frankell, A.M., Harrison, T. et al. Tracking early lung cancer metastatic dissemination in TRACERx using ctDNA. Nature 616, 553–562 (2023). https://doi.org/10.1038/s41586-023-05776-4
Martínez-Ruiz, C., Black, J.R.M., Puttick, C. et al. Genomic–transcriptomic evolution in lung cancer and metastasis. Nature 616, 543–552 (2023). https://doi.org/10.1038/s41586-023-05706-4